A critical initialization for biological neural networks
Neural recordings resemble linear dynamical systems governed by a symmetric connectivity matrix.
Abstract
Artificial neural networks learn faster if they are initialized well. Good initializations can generate high-dimensional macroscopic dynamics with long timescales. It is not known if biological neural networks have similar properties. Here we show that the eigenvalue spectrum and dynamical properties of large-scale neural recordings in mice (two-photon and electrophysiology) are similar to those produced by linear dynamics governed by a random symmetric matrix that is critically normalized. An exception was hippocampal area CA1: population activity in this area resembled an efficient, uncorrelated neural code, which may be optimized for information storage capacity. Global emergent activity modes persisted in simulations with sparse, clustered or spatial connectivity. We hypothesize that the spontaneous neural activity reflects a critical initialization of whole-brain neural circuits that is optimized for learning time-dependent tasks.
Thread:
- What if… spontaneous neural activity 🧠 reflects the baseline rumblings of a brainwide dynamical system initialized for learning? We find that the rumblings have macroscopic properties like those emerging from linear symmetric, critical systems 🧵 #neuroscience #neuroAI
- Long timescales and large principal components (PCs) can be produced by a dynamical system with random connectivity and independent stochastic inputs, if the connectivity matrix is critically-normalized.
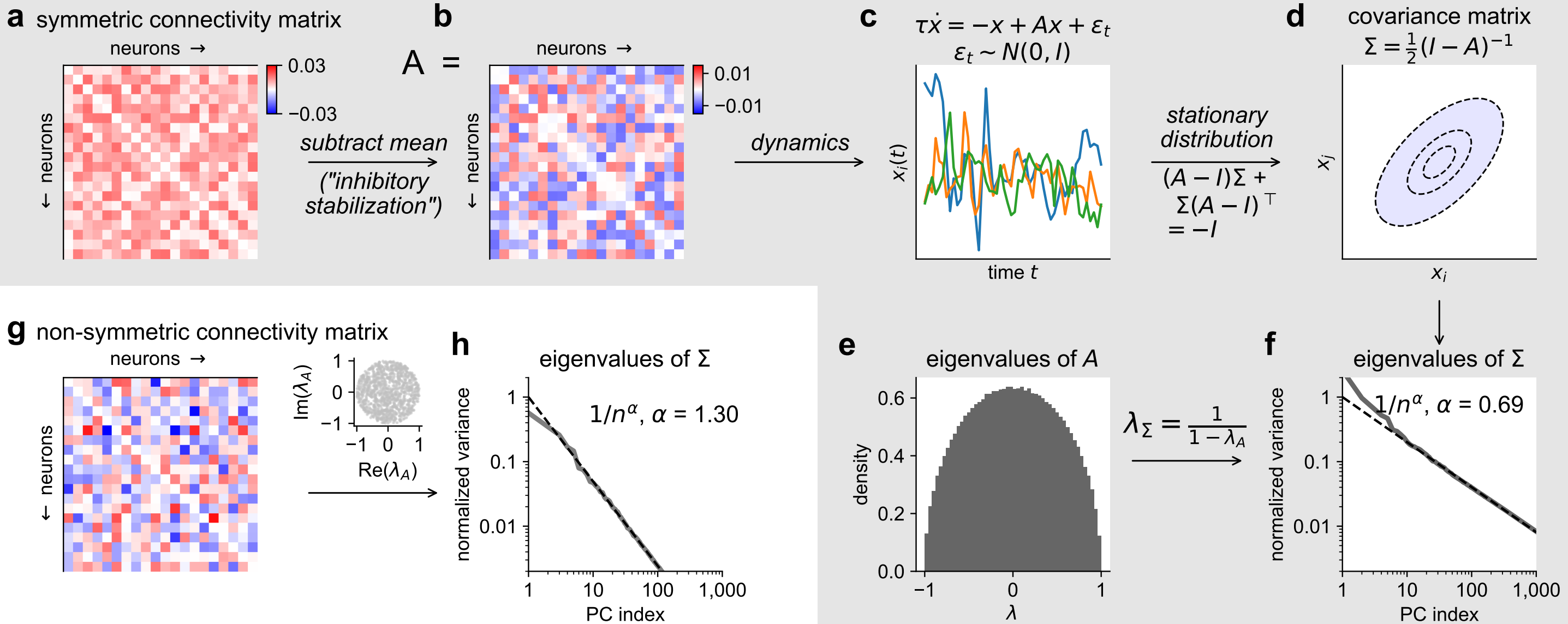
Furthermore, the principal components in the model decay as a power-law, a phenomenon we have previously reported in large-scale neural recordings: https://www.science.org/doi/10.1126/science.aav7893; https://www.nature.com/articles/s41586-019-1346-5
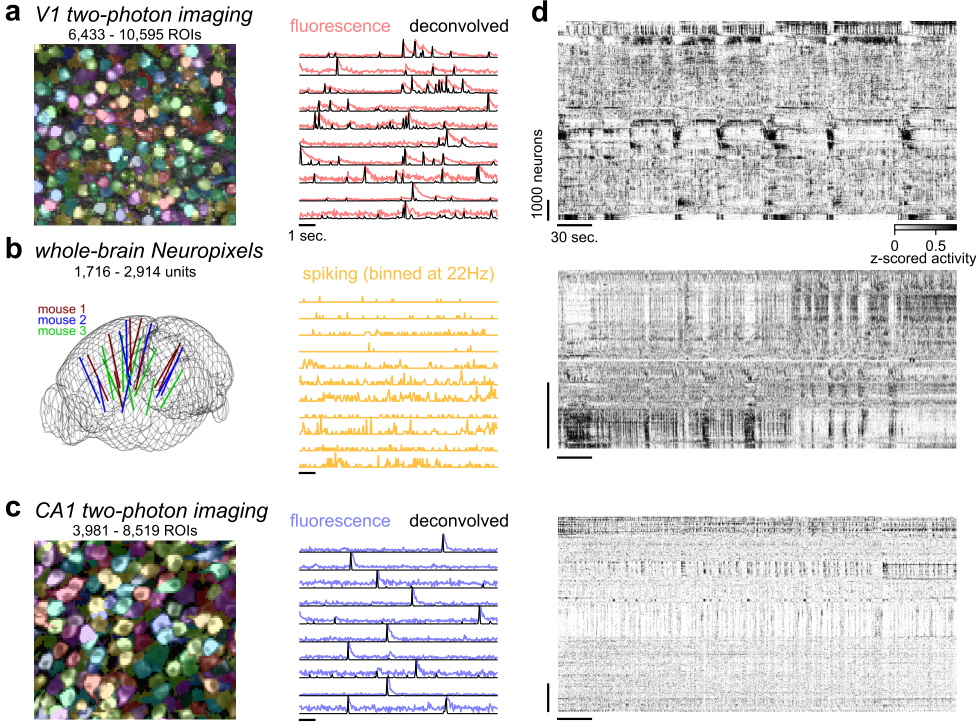
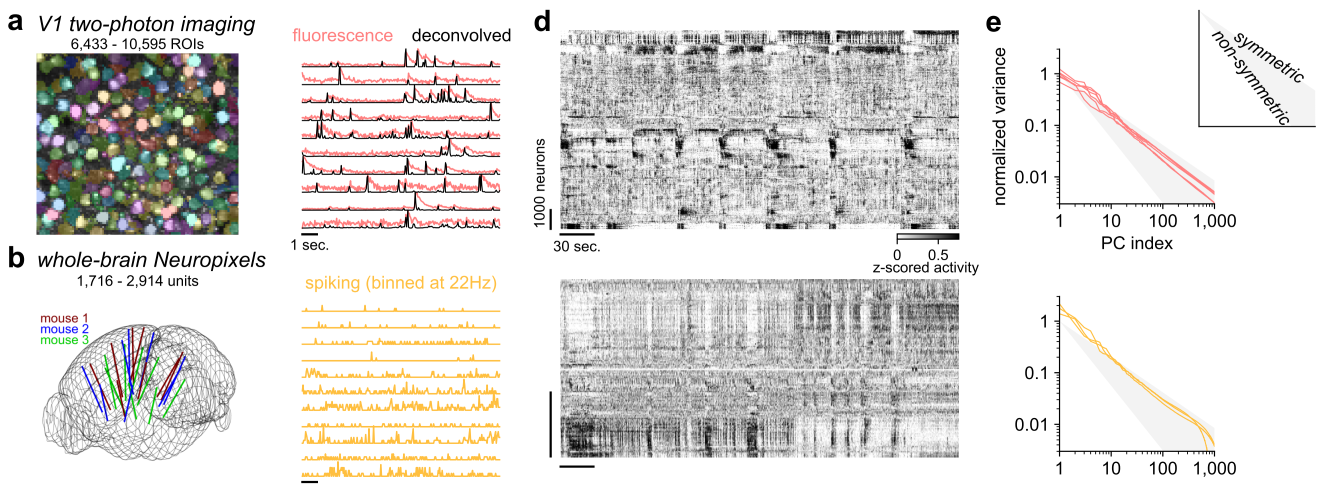
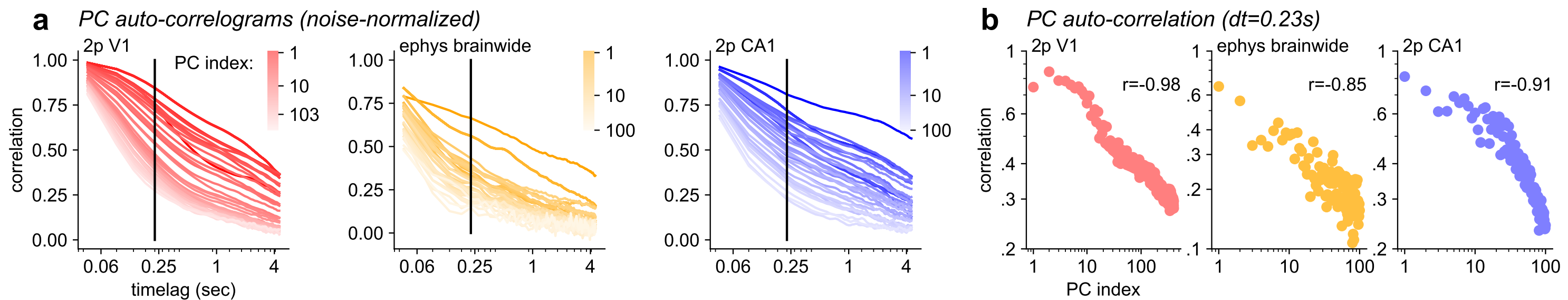
In V1 and brainwide ephys recordings we observed that the PC variances decayed as a power-law with exponents of 0.7-0.85, consistent with symmetric, critically-normalized simulations.
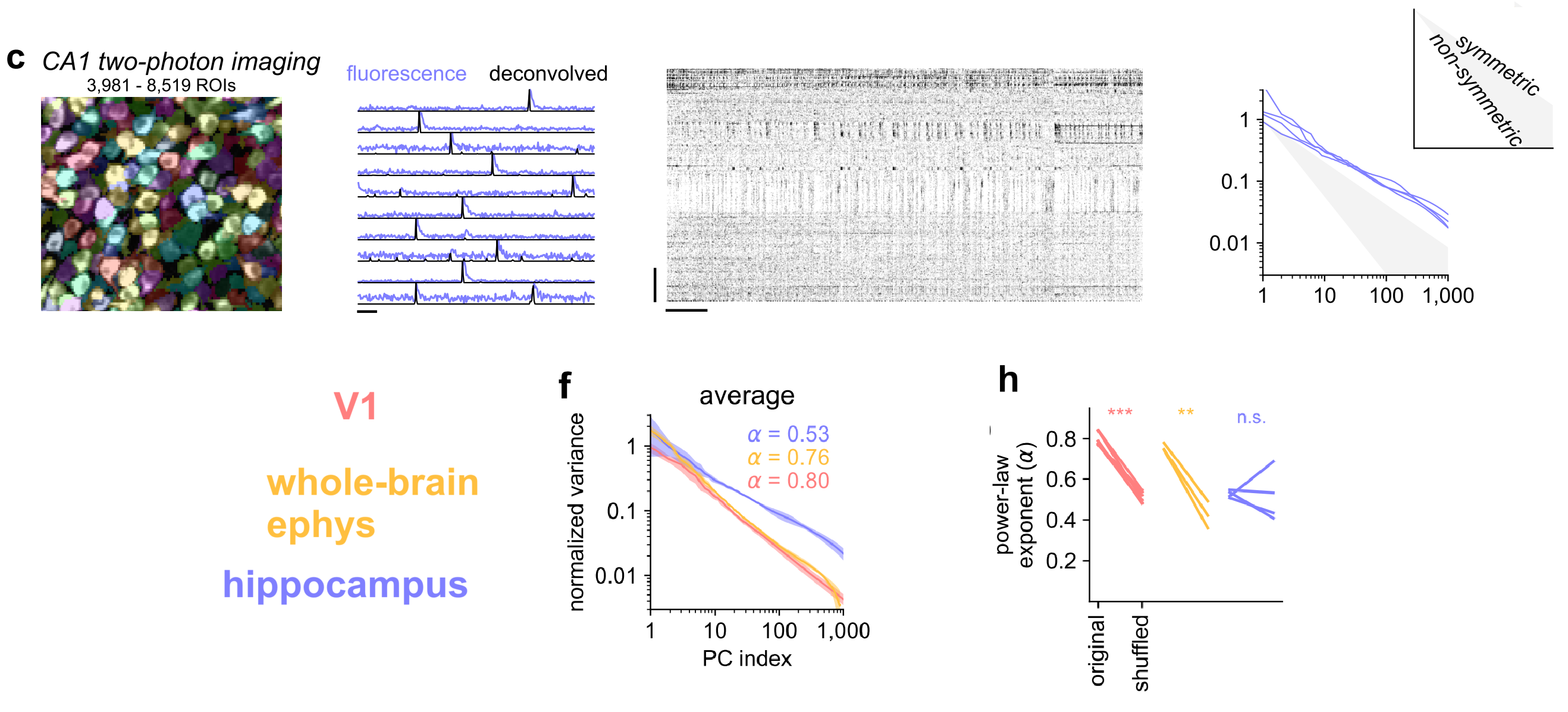
- But in hippocampus, we observed exponents around 0.5 that did not change after shuffling, suggesting that hippocampal activity is closer to completely independent neurons.
- The model also predicted that higher PCs have longer timescales, which was true in the data.
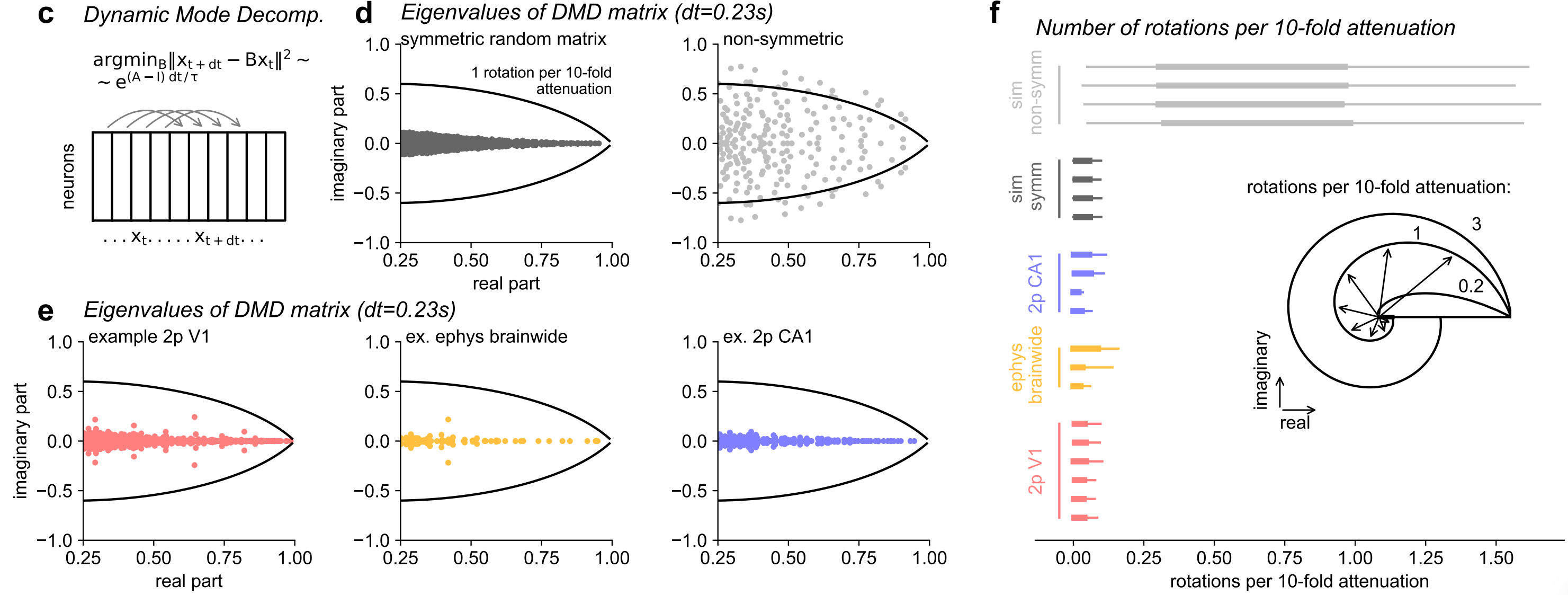
- An estimate of the dynamics matrix of the data (using DMD) revealed mostly real eigenvalues, further supporting symmetric dynamics.
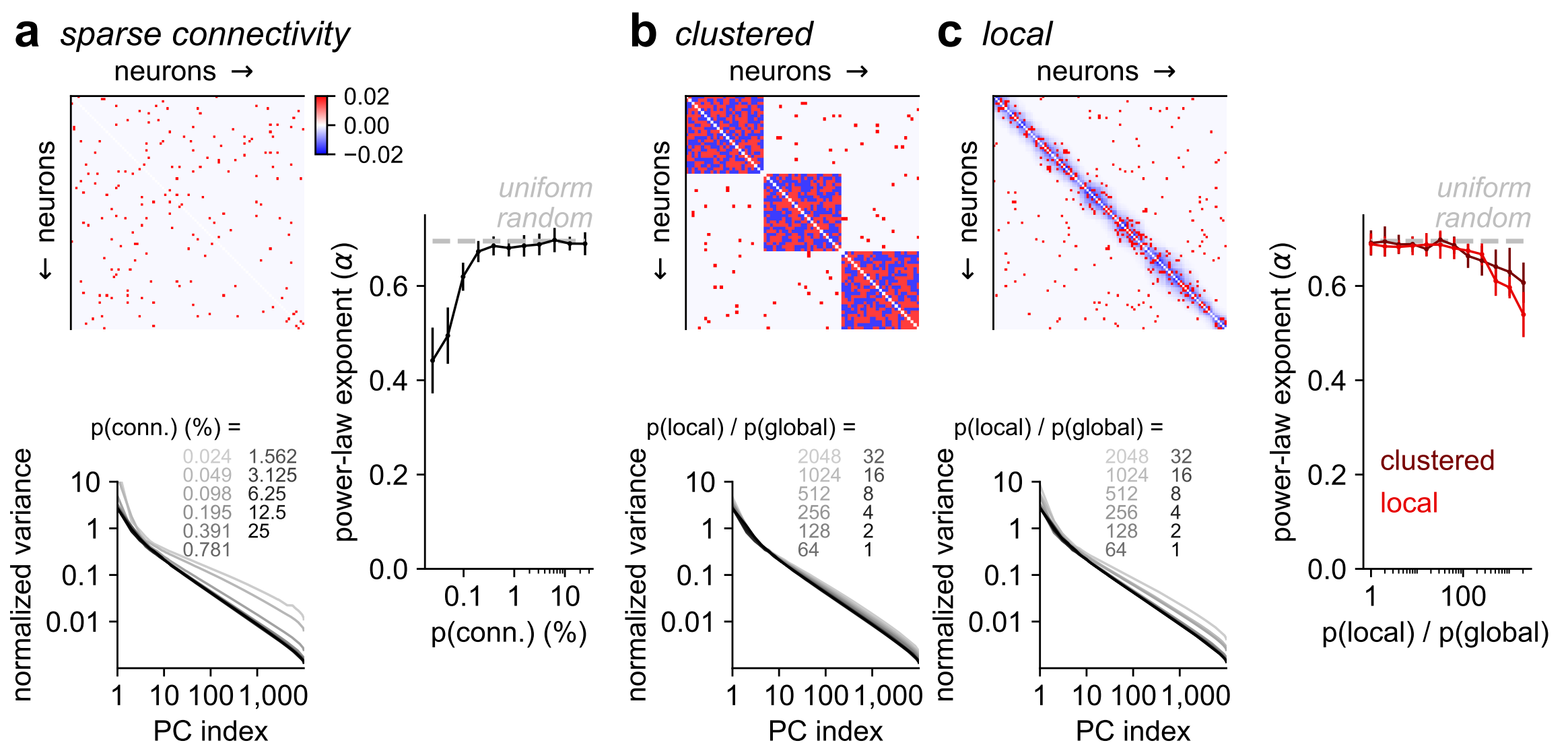
- Global emergent activity modes persisted in simulations with sparse, clustered or spatial connectivity.
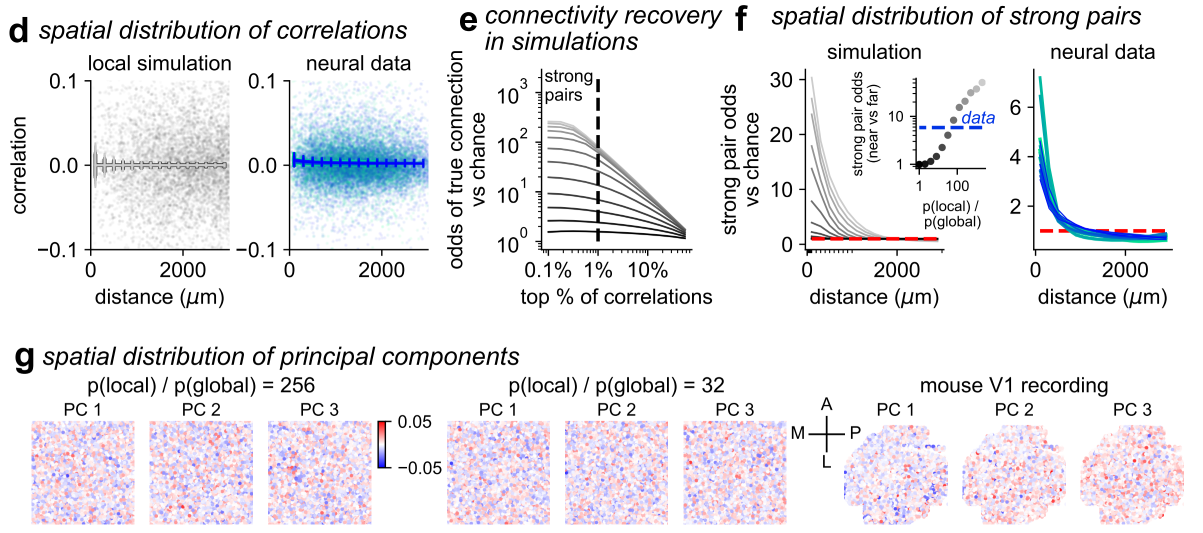
- Simulations with spatial connectivity replicated several of the properties we observed in the neural recordings, such as a spatial dependence of top correlated neuron pairs, and top PCs which were globally spread across cortex.
- We hypothesize that the spontaneous neural activity reflects a critical initialization of whole-brain neural circuits that is optimized for learning tasks that are time-dependent and working-memory dependent. More details in the paper by @marius10p.bsky.social.

Powered by Quarto. © Marius Pachitariu & Carsen Stringer lab, 2023.