Cellpose: a generalist algorithm for cellular segmentation
Accurate deep learning-based cellular segmentation tool that works for a wide variety of images, and includes an easy to use GUI.
Abstract
Many biological applications require the segmentation of cell bodies, membranes and nuclei from microscopy images. Deep learning has enabled great progress on this problem, but current methods are specialized for images that have large training datasets. Here we introduce a generalist, deep learning-based segmentation method called Cellpose, which can precisely segment cells from a wide range of image types and does not require model retraining or parameter adjustments. Cellpose was trained on a new dataset of highly varied images of cells, containing over 70,000 segmented objects. We also demonstrate a three-dimensional (3D) extension of Cellpose that reuses the two-dimensional (2D) model and does not require 3D-labeled data. To support community contributions to the training data, we developed software for manual labeling and for curation of the automated results. Periodically retraining the model on the community-contributed data will ensure that Cellpose improves constantly.
Thread:
- Releasing Cellpose, a generalist algorithm for cellular segmentation. Try it now directly on the website www.cellpose.org, or install the GUI with
pip install cellpose:
- We developed Cellpose as a generalist algorithm, because many small and big problems in biology require cell segmentation, and there just isn’t enough time to write a new pipeline for every type of data.

- We trained Cellpose on a diverse set of 608 images, collected and segmented by us. See the t-SNE plot of image styles below.
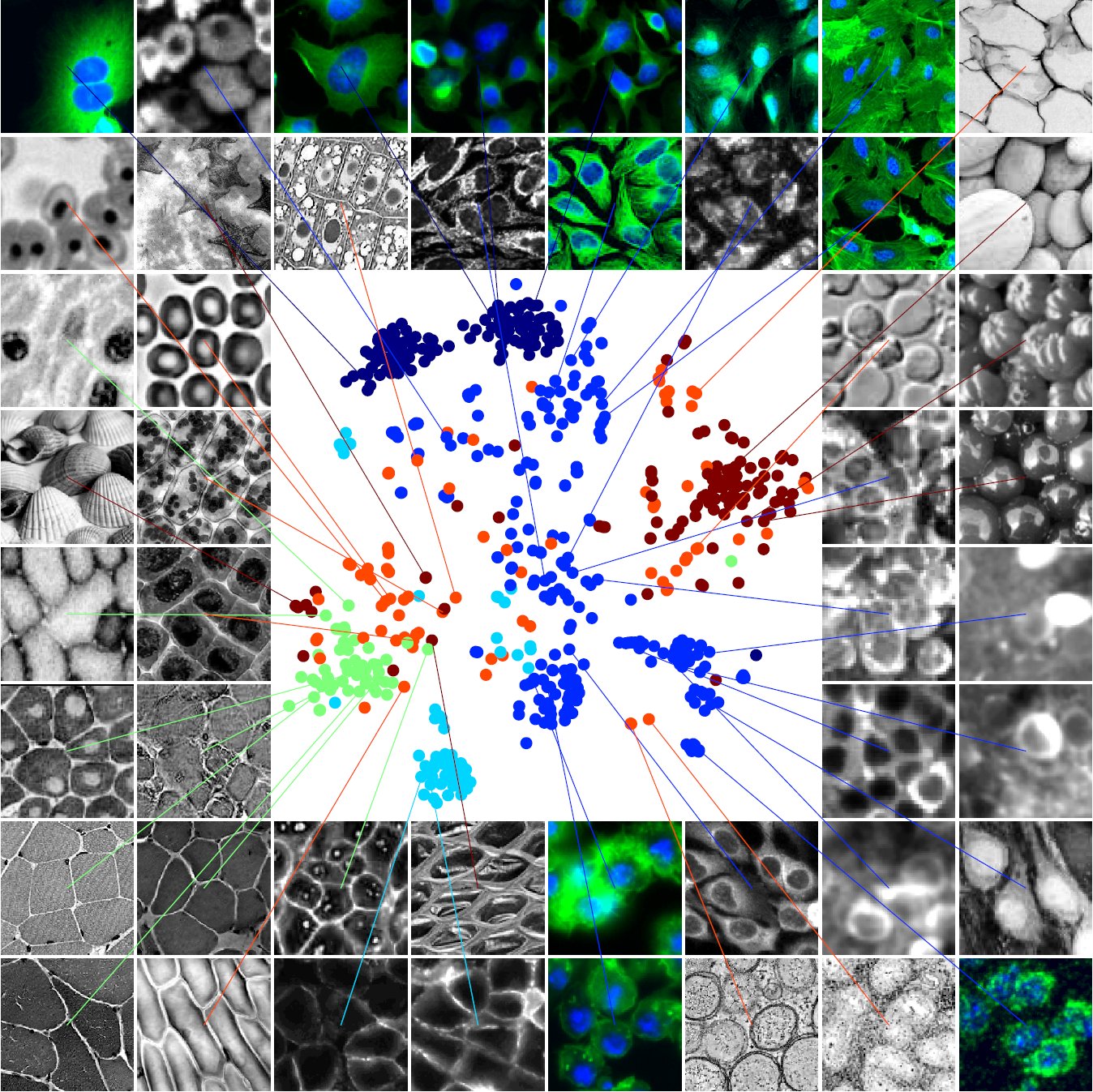
- Out-of-the box, Cellpose can segment a large variety of images from different types of microscopy, different tissues and different stains or fluorescent tags. It can even segment rocks, jellyfish and sea urchins.

- The GUI lets you manually segment your own images at a speed of 300-600 objects per hour. Cellpose doesn’t need super precise outlines.
- Send us your manual segmentations and we’ll include them in the next Cellpose release, making the model better for yourself and everyone else! P.S. thanks for sending us your segmentations, they are now in the “cyto2” model!

- Also you can perform 3D segmentation without 3D training data!


- Check out the [paper] for much more: nucleus segmentation, comparisons to previous state-of-the-art methods, a cell size prediction network etc.

Powered by Quarto. © Marius Pachitariu & Carsen Stringer lab, 2023.